LAB MATERIALS
R Markdown file for Lab 4 Click link to download. Fill it in with your answers to the following lab tasks. When you’re ready to submit, name it as
Lab4_FirstinitialYourlastname.Rmd, and submit it using the Sakai dropbox.Lab_4_kenya.rds - data file available on Sakai
Lab 4 Goals
Calculate measures of association between birth order (i.e. the primary exposure of interest) and child mortality by 5 years (i.e. the primary outcome of interest) within strata of other variables
Assess for potential confounding, mediation or effect measure modification
Lab 4 Grading scheme
| Competency | Points |
|---|---|
| .Rmd file runs without error | 10 |
| Table 1 - Frequency counts and Risk | 20 |
| Table 1 - Risk difference | 20 |
| Table 1 - Risk ratio | 20 |
| Task 3 (short answer) | 15 |
| Task 4 (short answer) | 15 |
| Total | 100 |
Task 1: Load libraries & data
For this assignment, use the dataset ‘Lab_4_kenya.rds’.
This lab will also require the packages {tidyverse}, {fmsb}, {epiR}, and {epiAssist}
You may need to install {fmsb}. You can do so with the following code:
install.packages("fmsb")
Hopefully at this point in the semester, you have a pretty good grasp on how to load libraries and data. But if you’re still unsure about something:
Help loading libraries. Help loading data.
Task 2: Table 1
INSTRUCTIONS: Generate frequency counts of 5-year
mortality (death) according to child’s birth order
(bord5), stratified by the following variables:
maleruralmageceducation
Use Table 1 to record frequency counts and strata-specific risk of
death before 60 months, by birth order. Then, use
epi.2by2() to generate stratified measures of risk
difference, risk ratios, and their accompanying confidence
intervals.
You will also use the resulting epi.2by2() object to
extract test statistics and p-values that test for potential effect
measure modification by the stratifying variables.
Moreover, you will use a pooled (i.e. adjusted) estimate of risk difference and risk ratio to assess for confounding by comparing those estimates to the crude (i.e. unadjusted) estimates of risk difference and risk ratio.”
Below is a detailed guide on how to complete each step of these instructions.
Tabular analysis of stratifying variables
Table 1 is an example of a stratified analysis of exposure and
outcome variables. Essentially, we want to know if the four variables
(male, rural, magec, and
education) are confounders and/or effect measure modifiers
of the relation between birth order and 5-year infant mortality. In
other words, Questions of effect measure modification include whether
the degree of association of child’s birth order and mortality status is
different for boys and girls? What about for rural and urban families?
Mothers of different age categories? And so on…
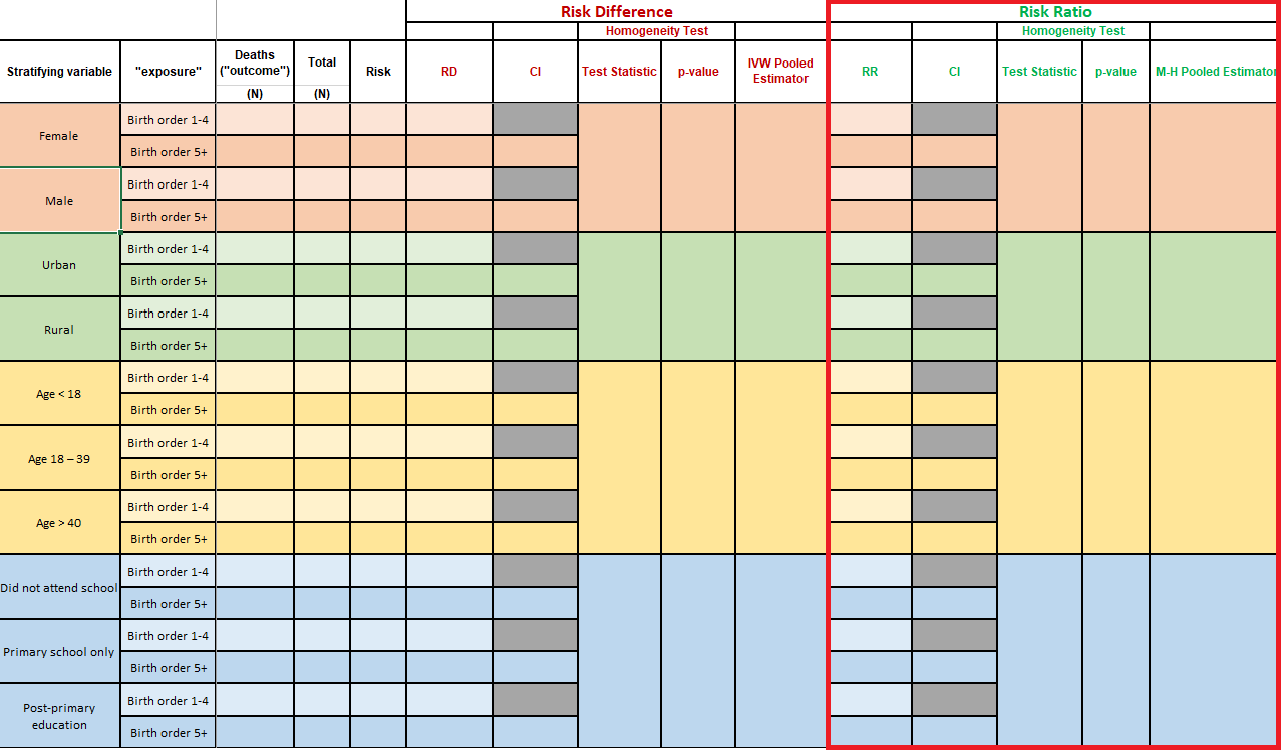
When you open the Excel file that contains Table 1, here is what you will see:
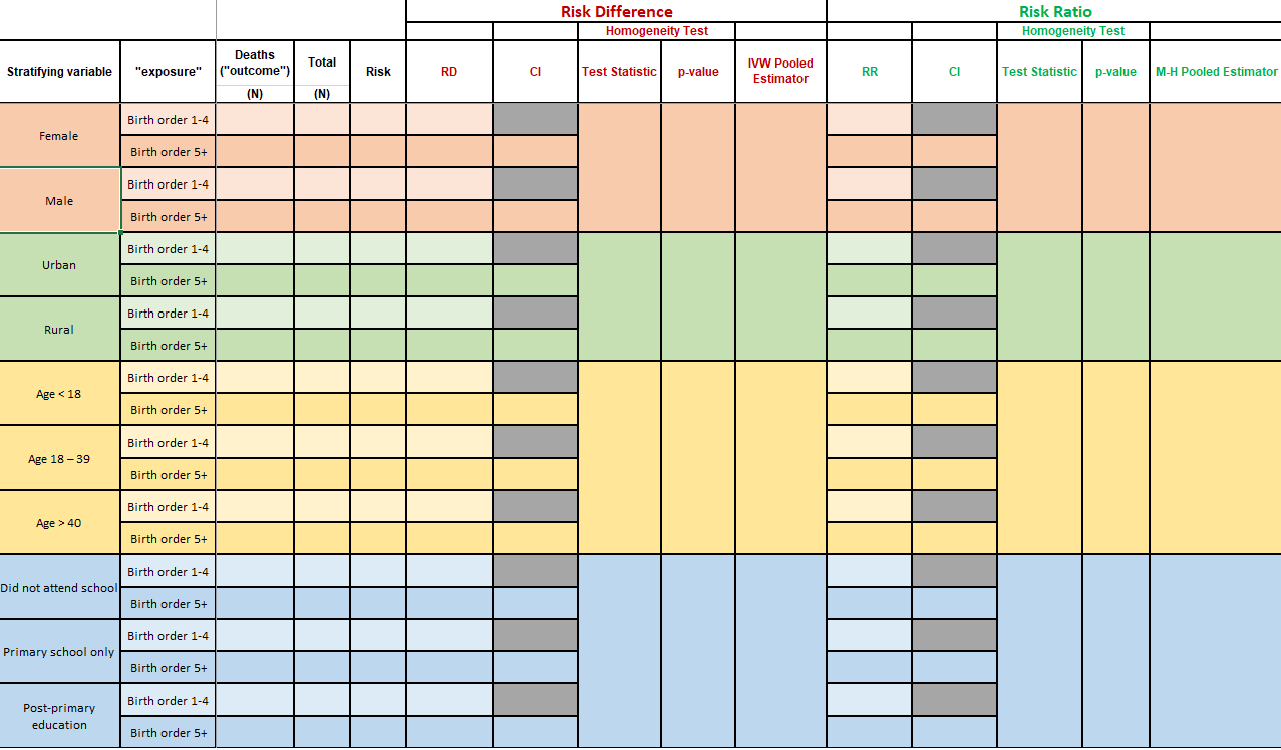
We will break Table 1 into sections, and walk you through how to obtain these values in R.
Task 2.1: Stratified frequency counts and risk
INSTRUCTIONS: Fill in the table with the numbers of
deaths (death, the outcome) and the 60-month risk of death
according to birth order (bord5, the exposure), stratified
by:
- Child’s gender (variable
male)
- Rural/urban residence (variable
rural)
- Maternal age (variable
magec)
- Maternal education (variable
education)
For this task, we recommend that you create a table object with
table(), similar to the
one you created in lab 3, where your outcome of interest is
mortality (death) and your exposure of interest is child
birth order (bord5). Except this time, include a third
stratifying variable.
table() allows for an unlimited number of stratifying
levels. You keep listing variables, and table() will keep
splitting the data into subsets.
Say that we created a binary variable in our kenya
dataset that indicated whether or not a mother’s BMI was above or below
25 kg/m2. Let’s call that variable mbmi_25. Here
is an example of a set of contingency tables of bord5 and
death, stratified by mbmi_25:
mbmi_tab <- table(kenya$bord5, kenya$death, kenya$mbmi_25)
mbmi_tab
#> , , = <25
#>
#>
#> Alive Dead
#> 1-4 in Birth Order 7931 1071
#> 5+ in Birth Order 2180 409
#>
#> , , = >=25
#>
#>
#> Alive Dead
#> 1-4 in Birth Order 3700 374
#> 5+ in Birth Order 773 146
Here, our stratifying variable is mbmi_25, which
separates our tabulated data into two tables according to mothers who
have a BMI greater than or equal to 25, and those with BMI less than
25.
We can generate row-wise proportions of our table by wrapping the
table object in the proportions() function.
proportions(mbmi_tab, margin = c(1, 3))
#> , , = <25
#>
#>
#> Alive Dead
#> 1-4 in Birth Order 0.88102644 0.11897356
#> 5+ in Birth Order 0.84202395 0.15797605
#>
#> , , = >=25
#>
#>
#> Alive Dead
#> 1-4 in Birth Order 0.90819833 0.09180167
#> 5+ in Birth Order 0.84113166 0.15886834
Task 2.2: Create
epi.2by2() object
INSTRUCTIONS: Create an epi.2by2()
object for each of the tables you generated above.
epi.2by2() works just like mAssoc(). We
need to rearrange
our tables so that our outcome of interest and exposure of interest
are in the top-left corner of each table.
Good news: The function flipTable() is designed to work
with stratified tables too!
Do the following:
a. flipTable() so that the cell that
contains the index level of the exposure (birth order) and the outcome
of interest (mortality status) is in the top-left corner of either
table.
For instance:
mbmi_flip <- flipTable(mbmi_tab)
b. Create an epi.2by2() object by using
the flipTable() object, with confidence level of 0.95 and
units set to 1.
mbmi_epi <- epi.2by2(mbmi_flip, units = 1, conf.level = 0.95)
Task 2.3: Extract estimates of risk difference
INSTRUCTIONS: Use the epi.2by2() object
to extract the measure of risk difference for mortality in association
with bord5 within each covariable stratum, along
with test statistics and an overall (“adjusted”) pooled estimate of risk
difference.
IMPORTANT: The output for epi.2by() is
NOT the full extent of the information contained within an
epi.2by2() object! There is more to every object than what
meets the eye.
When you call an epi.2by2() object, you will notice that
its printed output is a lot like that of mAssoc() from the
previous lab. Except this time, there are “crude” and “M-H”
estimates:
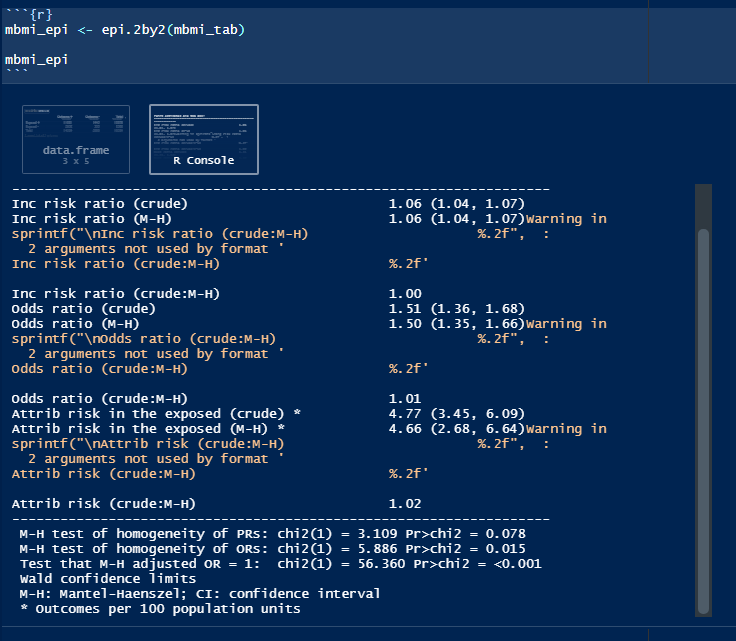
But if you inspect the epi.2by2() object (either in your
R Environment, or by typing the object’s name, followed by
$), you will find vectors, lists, data frames, and all
sorts of sub-objects.
Following is how to locate the necessary information for this portion of Table 1
a. Risk difference and 95% CI
To access the risk differences of individual strata, use the
following path to refer to that exact component of your
epi.2by2 object:
object$massoc.detail$ARisk.strata.wald
So to obtain our measures of risk difference within separate stratum of mothers with BMI < 25 and mothers with BMI >= 25, we can use the following code:
mbmi_epi$massoc.detail$ARisk.strata.wald
#> est lower upper
#> 1 0.03900249 0.02344295 0.05456203
#> 2 0.06706667 0.04182403 0.09230931
You might notice that this returns a single estimate for each stratum. Our table, however, asks for two estimates for each. What gives??
Since we have pre-determined our “reference” and “index” levels, and know that a risk difference is defined as “risk in the level of interest minus risk in the reference level”, what’s the result when our reference level is also our level of interest?
Check your work!
To check which value belongs to which strata, you can calculate it by hand using the values for risk that you obtained in Task 2.1.
You can also use the function riskdifference() from
package {fmsb} to quickly derive risk ratios and confidence
intervals.
To learn how to use this function, you can view its documentation by running the following code in your console:
?riskdifference
b. Test of homogeneity and p-value
Inconveniently, epi.2by2() doesn’t actually provide a
chi-squared test of homogeneity of stratified risk differences. We can
obtain this estimate by using the function epiHomog(),
which takes a flipTable() object as its primary
argument.
If your flipTable() objects are properly oriented, they
should suffice. Use your flipTable() objects as an argument
in the function epiHomog() to return the test statistic and
p-value for a test of homogeneity at a significance level of 0.05:
epiHomog(aFlippedTable)
Using our flipTable() object that is stratified by
mother’s BMI, we would run the following code to obtain the resulting
output:
epiHomog(mbmi_flip)
#> A tibble: 1 x 2
#> `Test statistic` `p-value`
#> <dbl> <dbl>
#> 1 3.44 0.0637
Interpreting X2 test of homogeneity
What exactly is the chi-square test of homogeneity for? Well, it’s all about effect measure modification. Suppose we focus on the risk difference measure. We use the risk difference measure to quantify the association of birth order with the risk of death by 60 months of age. In particular, we use it to quantify the risk of death for those with birth order 5+ in comparison to the risk of death for those with birth order 1-4 (i.e. the reference level) by subtracting the two risks.
When we consider a third variable such as male, the chi-square test of homogeneity is used to evaluate whether there is evidence that the risk difference is different for male and for female children. If so, it is important that we report the risk difference separately for male and for female children. In an interesting feature, if there is effect measure modification on one effect measure scale (e.g. risk difference), then it may very well not be present on another scale (e.g. risk ratio). This is precisely why we talk about “effect measure modification” and not simply “effect modification”.
c. Pooled estimate (Mantel-Haenszel)
Finally, we want the pooled estimate of risk difference. You might
have already found this in the original print method’s output of our
epi.2by2() object, where it’s referred to as
Attrib risk in the exposed (M-H).
But since we need to report this estimate to three decimal points
(see gutter), we need to reach into our epi.2by2 object and
find the exact Mantel-Haenszel pooled estimate. You can access it by
calling the following path:
object$massoc.detail$ARisk.mh.wald
Using our epi.2by2() object from before,
mbmi_epi, we find this value and its resulting output as
follows:
mbmi_epi$massoc.detail$ARisk.mh.wald
#> est lower upper
#> 1 0.04662555 0.02684395 0.06640716
We would interpret this pooled result by saying that, when accounting
for confounding in the exposure due to a third variable
(mbmi_25), risk of the outcome in the exposed was 0.05 (95%
CI 0.03 to 0.07) higher than the unexposed.
Task 2.4: Extract estimates of risk ratios
The process for obtaining risk ratios, test statistics for
homogeneity between strata, and pooled estimates is very similar to that
of obtaining risk differences. Except this time, we don’t need a
separate function for finding the test of homogeneity –
epi.2by2() generates the test statistic for homogeneity of
risk ratios itself.
Risk ratios
To extract risk ratios from an epi.2by2() object, you
can use the following path:
object$massoc.detail$RR.strata.wald
Again, when you’re finding these risk ratios, consider what we’re asking for in the table when we request the risk ratio of the reference level as it pertains to the reference level. What does it amount to when we take the ratio of something to itself?
](https://imgs.xkcd.com/comics/honor_societies.png)
Figure 2: Tautology club
Okay moving on…so our mbmi_epi object renders the
following output:
mbmi_epi$massoc.detail$RR.strata.wald
#> est lower upper
#> 1 1.046320 1.027315 1.065676
#> 2 1.079734 1.048089 1.112334
Check your work!
To check which value belongs to which strata, you can calculate it by hand using the values for risk that you obtained in Task 2.1.
You can also use the function riskratio() from package
{fmsb} to quickly derive risk ratios and confidence intervals.
To learn how to use this function, you can view its documentation by running the following code in your console:
?riskratio
Test of homogeneity and p value
The test of homogeneity for risk ratios is available in the following
location within our epi.2by2() object
object$massoc.detail$wRR.homog
Performing this on our mbmi_epi object yields the
following result:
mbmi_epi$massoc.detail$wRR.homog
#> test.statistic df p.value
#> 1 3.109451 1 0.0778392
Reflecting on this test statistic and the resulting p-value, given a significance level of 0.05, would we conclude that mother’s BMI is a confounder in assessing the influence of birth order on the 5 year risk of mortality of children in our study cohort?
Hover for answer: YOU THINK YOU GET FREEBIES??
Pooled estimate (Mantel-Haenszel)
The pooled estimate appears in the printed output when you call your
epi.2by2() object from the environment, and is called
Inc risk ratio (M-H). However, this is only reported to two
decimals. To access the actual value instead of the rounded one, you can
look for it in the following place within your epi.2by2()
object:
object$massoc.detail$RR.mh.wald
Using our mbmi_epi object renders the following
output:
mbmi_epi$massoc.detail$RR.mh.wald
#> est lower upper
#> 1 1.055389 1.039048 1.071988
We would interpret this as meaning that when we account for confounding due to mother’s BMI, the risk of experiencing the outcome among the exposed was 1.06 (95% CI 1.04 to 1.07) times greater than the outcome of experiencing risk among the unexposed.
Task 3: Short Answer
PROMPT: Do you see evidence for confounding for any
of the stratification variables (male, rural,
magec, and education)? Explain your response
for each variable.
(Hint, you will need to refer back to the crude association estimates of risk difference and risk ratio that you generated for Table 3 of Lab 3).
Task 4: Short Answer
PROMPT: For which covariables does it seem appropriate to report a single pooled adjusted estimate and for which does it seem necessary to report separate stratified estimates (i.e., do you see evidence of effect modification)? Explain your rationale for each covariable.
Does your answer differ depending on whether you are considering the risk difference or risk ratio as your measure of effect?