LAB MATERIALS
R Markdown file for Lab 1 Click link to download. Fill it in with your answers to the following lab tasks. When you’re ready to submit, name it as
Lab1_FirstinitialYourlastname.Rmd, and submit it using the Sakai dropbox.Lab_1_kenya.rds - data file available on Sakai
Lab 1 Goals
- Generate derived variables
- Identify and recode special values
- Run descriptive statistics for continuous and categorical variables
- Generate graphics for continuous variables
- Generate a complete data dictionary
Lab 2 Grading scheme
| Competency | Points |
|---|---|
| .Rmd file runs without error | 10 |
| Task 3 (Histograms) | 15 (5 each) |
| Task 4 (Boxplots) | 15 (5 each) |
| Task 5 (Table 1) | 10 |
| Task 6 (Data Dictionary - original variables) | 20 |
| Task 6 (Data Dictionary - new variables) | 10 |
| Task 7 (short answer) | 20 |
| Total | 100 |
Task 1: Load packages and data
In the chunk of code named load packages, use the
function library() to load packages {skimr} and
{tidyverse}.
In the following chunk, named load data, use the
function readRDS() to load in the dataset named
Lab_1_kenya.rds. Use the assignment operator,
<- to give it the name kenya.
Reminder: To run a line of code from within
a code chunk in R Markdown, use the keyboard shortcut
ctrl + enter (Windows) / cmd + enter (Mac)
Task 2: Recode variables
Similar to Lab 0, construct the following derived variables (Tasks 2a
- 2e) using a pipe (%>%) and mutate(). For
each, examine the component variables for coded special values
(i.e. “NA”) and be sure to set derived variable values appropriately.
For categorical variables, you will be expected to use
case_when() to create conditional clauses, just as you did
in Lab 0 to
derive magec.
For the categorical variables: Label all variables
and the coded values for the categorical ones using the
factor() command. That is, add meaningful labels to both
the variable name itself and to the levels (i.e. categories) of the
categorical variable to make printed output more readable.
In the .Rmd named lab1_705_fall2021.Rmd, we have provided you with an
example of the correct coding of bord5. Please use this to
assist you as you re-code the other variables that follow. Please code
each new variable within the code chunk named after that variable.
Task2a: bord5
bord5: A dichotomous categorical
(i.e. binary) variable indicating birth order of the child. You will use
the variable bord (birth order). Create the categories for
bord5 according to the following levels and labels:
| Level | Label |
|---|---|
| 0 | birth order 1 through 4 |
| 1 | birth order 5+ |
Task 2b: male
male: A dichotomous categorical
(i.e. binary) variable indicating that the child is male. Based on
variable b4. Create the categories for male
according to the following levels and labels:
| Level | Label |
|---|---|
| 0 | female |
| 1 | male |
Task 2c:
mweight
mweight: Continuous variable for
maternal weight at time of interview (in kilograms). Based on variable
v437. Note that v437 contains 1 implied
decimal place. Divide by 10 to get kilograms.
Task 2d:
mheight
mheight: Continuous maternal height at
time of interview (in meters). Based on variable v438. Note
that this variable is in centimeters and also contains 1 implied decimal
place. Divide by 10 to get meters.
Task 2e: mbmi
mbmi: maternal body mass index (BMI).
mweight / mheight^2.
Task 3: Frequency histograms
Generate frequency histograms of the continuous variables
mweight,
mheight and
mbmi using {ggplot2}. Put meaningful axis
labels and a title on each figure. Note that since we look at histograms
of each of the variables individually, these are univariable
visualizations (i.e. one variable at a time).
Please read the description below on how to generate histograms in
{ggplot}, then use the code chunks named mweight hist,
mheight hist, and mbmi hist to generate each
plot in its own designated chunk.
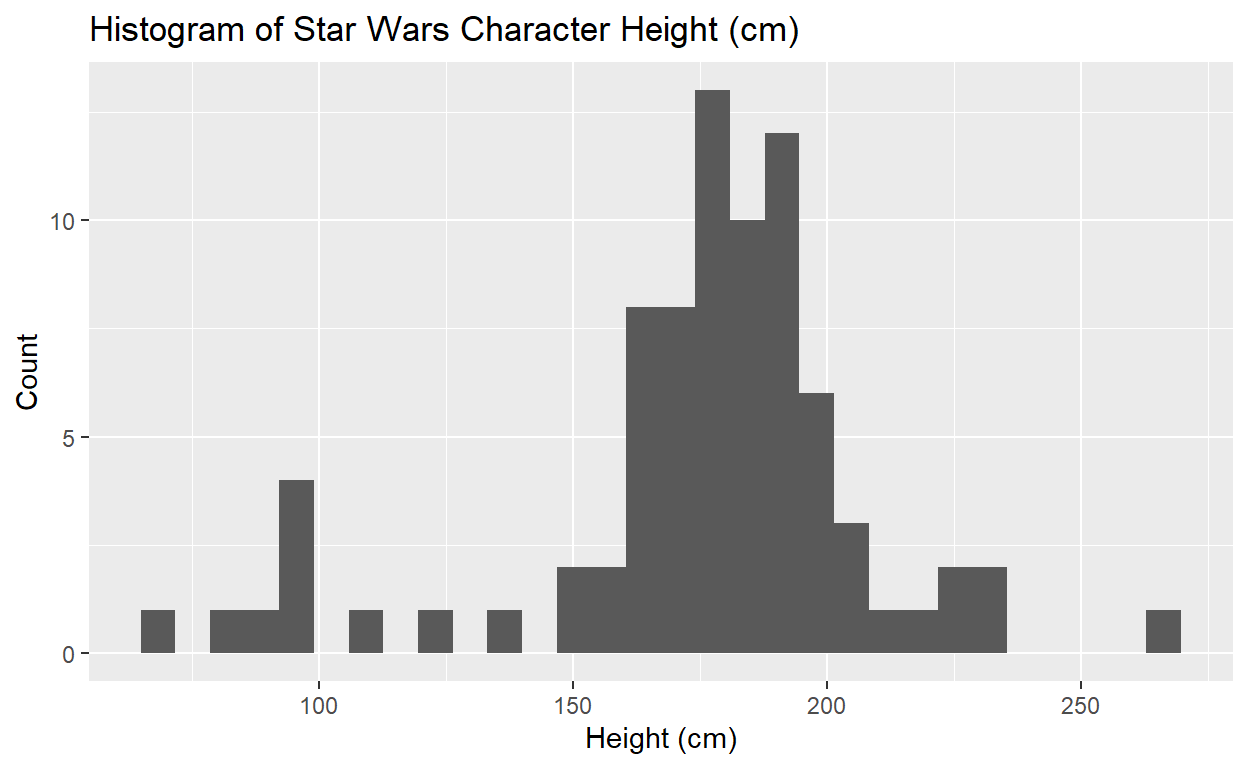
Creating histograms in {ggplot2}
The code for creating histograms using ggplot is fairly
straightforward, and we will demonstrate using the starwars
dataset. It provides a list of Star Wars characters, with details on
their height, weight, gender, home world, and so on:
| name | height | mass | hair_color | skin_color | eye_color | birth_year | sex | gender | homeworld | species |
|---|---|---|---|---|---|---|---|---|---|---|
| Luke Skywalker | 172 | 77 | blond | fair | blue | 19.0 | male | masculine | Tatooine | Human |
| C-3PO | 167 | 75 | NA | gold | yellow | 112.0 | none | masculine | Tatooine | Droid |
| R2-D2 | 96 | 32 | NA | white, blue | red | 33.0 | none | masculine | Naboo | Droid |
| Darth Vader | 202 | 136 | none | white | yellow | 41.9 | male | masculine | Tatooine | Human |
| Leia Organa | 150 | 49 | brown | light | brown | 19.0 | female | feminine | Alderaan | Human |
| Owen Lars | 178 | 120 | brown, grey | light | blue | 52.0 | male | masculine | Tatooine | Human |
Say we want to use ggplot to create a histogram of
character height. Where do we start?
“gg” stands for Grammar of graphics. {ggplot2} allows us to follow simple rules, a “grammar”, to construct visually appealing graphics and plots.
Regardless of what kind of visualization we want to create, the first
step will always be to initialize a graphical field using the command
ggplot(). We supply this function with our dataset using
the argument data =.
We can also supply it with universal “mapping aesthetics”. What does
that mean? Generally speaking, “aesthetics” are the different
variables we want to use in our plot. Eventually, we are going to add
“layers” to the plot. A universal aesthetic just means that we
want the variable to apply to ALL layers. In the case of our histogram,
we want height to always be on the x-axis. So we will
designate height as a universal aesthetic. We designate
aesthetics by putting them inside the aesthetic function,
aes().
Notice how the above plotting field is initialized, with character
height mapped to the x axis. Luckily,
histograms only take an x aesthetic, so we can generate our histogram by
simply adding a layer using geom_histogram(). {ggplot2}
uses a plus sign, + to add layers
to a plot. Here, we will add a histogram layer and a labels layer
(labs())
ggplot(data = starwars, mapping = aes(x = height)) +
geom_histogram() +
labs(x = "Height (cm)", y = "Count", title = "Histogram of Star Wars Character Height (cm)")
Please pay special attention to how you label your visualizations, and be specific! Statisticians and others reviewing your research tend to be quite persnickety when it comes to titles and axis labels. And with good reason! What use is a visualization without proper labels?
As time permits, we encourage you to take advantage of the external resources provided in the sidebar in order to gain a better understanding of plotting with {ggplot2}
Task 4: Boxplots
Now you’re going to generate boxplots of mweight,
mheight, and mbmi for the levels of
magec. Put meaningful axis labels and a title on each
figure. Note that since we look at boxplots of each of the variables
according to the levels of magec, these are bivariable
visualizations (i.e. showing two variables at a time).
Please use the appropriately labelled code chunks provided in your R
Markdown assignment document (labelled mweight box,
mheight box, etc.) to generate your boxplots.
Creating a boxplot in {ggplot2}
The steps to creating a boxplot are the same as those for creating a
histogram, except we will create the boxplot layer using
geom_boxplot() instead of geom_histogram().
Additionally, in order to stratify our plot, we will need to provide the
plotting space with a y aesthetic, magec.
If you need a little more guidance, learn to create a boxplot here
And learn how to create a stratified boxplot here
Task 5: Frequency table (Table 1)
If you haven’t already, download the Excel file that’s linked at the top of this webpage. Then
open the file, which is named Lab1_Tables.xlsx.
Fill in the sheet called Table 1 with the frequency counts and percentages for the levels of the 3 categorical variables you have generated. Calculate percentages only for the non-missing values. Round percentages to 1 decimal place.
In the code chunks named bord5 freq, etc., we would like
you to use a pipe (%>%) with group_by(),
summarize(), and mutate() to generate summary
statistics. Please read the tutorial below to gain a better
understanding of group_by() and
summarize().
Cross-tabulation
with group_by() and summarize()
group_by() works by grouping rows into discrete
categories based on categorical variables in the data frame. It then
allows a function like summarize() to calculate summary
statistics on those sub-groups, including percentages and frequency
counts.
As an example, we can use the mtcars dataset, which is
always available from within R:
head(mtcars)
| mpg | cyl | disp | hp | drat | wt | qsec | vs | am | gear | carb | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mazda RX4 | 21.0 | 6 | 160 | 110 | 3.90 | 2.620 | 16.46 | 0 | 1 | 4 | 4 |
| Mazda RX4 Wag | 21.0 | 6 | 160 | 110 | 3.90 | 2.875 | 17.02 | 0 | 1 | 4 | 4 |
| Datsun 710 | 22.8 | 4 | 108 | 93 | 3.85 | 2.320 | 18.61 | 1 | 1 | 4 | 1 |
| Hornet 4 Drive | 21.4 | 6 | 258 | 110 | 3.08 | 3.215 | 19.44 | 1 | 0 | 3 | 1 |
| Hornet Sportabout | 18.7 | 8 | 360 | 175 | 3.15 | 3.440 | 17.02 | 0 | 0 | 3 | 2 |
| Valiant | 18.1 | 6 | 225 | 105 | 2.76 | 3.460 | 20.22 | 1 | 0 | 3 | 1 |
Say we want to know the frequency counts of cars with 4, 6, and 8
cylinders (variable: cyl) based on whether or not a car has
an automatic or manual transmission (variable am, 0 =
automatic, 1 = manual).
We can use group_by() to create subgroups according to
am and cyl. Using group_by() by
itself doesn’t alter the appearance of our data frame, but our
observations will now be grouped implicitly according to transmission
type and number of cylinders.
| mpg | cyl | disp | hp | drat | wt | qsec | vs | am | gear | carb |
|---|---|---|---|---|---|---|---|---|---|---|
| 21.0 | 6 | 160 | 110 | 3.90 | 2.620 | 16.46 | 0 | 1 | 4 | 4 |
| 21.0 | 6 | 160 | 110 | 3.90 | 2.875 | 17.02 | 0 | 1 | 4 | 4 |
| 22.8 | 4 | 108 | 93 | 3.85 | 2.320 | 18.61 | 1 | 1 | 4 | 1 |
| 21.4 | 6 | 258 | 110 | 3.08 | 3.215 | 19.44 | 1 | 0 | 3 | 1 |
| 18.7 | 8 | 360 | 175 | 3.15 | 3.440 | 17.02 | 0 | 0 | 3 | 2 |
| 18.1 | 6 | 225 | 105 | 2.76 | 3.460 | 20.22 | 1 | 0 | 3 | 1 |
We might use summarize() to operate on these sub-groups.
summarize() works with the following syntax:
Depending on whether our variable of interest is categorical or
continuous, we can use summary statistic functions like n()
(for raw counts), mean(), sd(),
median(), IQR(), min(), and
max() within our summarize() function. Except
for certain functions like n(), which doesn’t take any
arguments, the rest of these functions take a variable name specifying
which variable should be used in the calculation.
We can start by using n() for sub-group frequency
counts. Notice that, similar to the syntax used with
mutate(), we name the summary statistic to the left of the
equals sign, and specify the function to the right.
| am | cyl | Counts |
|---|---|---|
| 0 | 4 | 3 |
| 0 | 6 | 4 |
| 0 | 8 | 12 |
| 1 | 4 | 8 |
| 1 | 6 | 3 |
| 1 | 8 | 2 |
We can also use mutate() to operate on our summary table
as if it were its own data frame.
Within mutate(), we create a new variable with
NewVariable =, and equate it to the following operation:
Counts / sum(Counts, na.rm = TRUE) (sub-group counts
divided by the sum of sub-group counts).
| am | cyl | Counts | Proportions |
|---|---|---|---|
| 0 | 4 | 3 | 0.1578947 |
| 0 | 6 | 4 | 0.2105263 |
| 0 | 8 | 12 | 0.6315789 |
| 1 | 4 | 8 | 0.6153846 |
| 1 | 6 | 3 | 0.2307692 |
| 1 | 8 | 2 | 0.1538462 |
Also notice that we’re able to do this all within a single string of
code using %>%!
Task 6: Data dictionary (Table 2)
In your Excel spreadsheet, the sheet titled Table 2
is our own data dictionary for the Kenya DHS dataset, with columns added
to annotate the variables and provide summary statistics. Using output
from skim():
- Fill in the columns in this table for the variables originally in the dataset.
- Add rows for the 7 new variables that you have created in both Labs 0 and 1.
Task 7: Short answer
Enter your response into your own RMarkdown file, under the heading called “Task 7: Short answer”
Prompt: Examine the range and proportion of missing values for each of the 7 variables you have created in Labs 0 and 1. Are there characteristics of any of these variables that are concerning (e.g., missing, suspicious or impossible values)? In contemplating analysis of these data, what do you think should be done with anomalous information? What effect would missing values have on the validity of your analyses (e.g., how might missing or extreme values affect inferences)? (Response no longer than 250 words, please)